Dimensionality Reduction
For
- Data Compression
- Data Visualization
Principal Component Analysis (PCA)
- Reduce from n-dimension to k-dimension:
1. Compute “covariance matrix”
2.Compute “eigenvectors” of matrix $\Sigma$
[U,S,V] = svd(Sigma);
we get:
reduce dimensionality:
Ureduce = U(:,1:k);
z = Ureduce'*x;
Chooosing $k$ (number of principal components)
- Average squared projection error: $\frac{1}{m} \sum_{i=1}^m | x^{(i)} - x_{approx}^{(i)} |^2 $
-
Total variation in the data: $\frac{1}{m} \sum_{i=1}^m | x^{(i)} |^2$
- Typically, choose $k$ to be smallest value so that:
99% of varianve is retained
Algorithm
[U,S,V] = svd(Sigma);
Pick smallest value of $k$ for which
99% of varianve is retained
Reconstraction from Compressed Representation
we know:
reconstraction:
Addvice for Applying PCA
Supervised learning speedup
$(x^{(1)},y^{(1)}),(x^{(2)},y^{(2)}),\dots,(x^{(m)},y^{(m)})$
Extract inputs: Unlabeled dataset:
New training set:
$(z^{(1)},y^{(1)}),(z^{(2)},y^{(2)}),\dots,(z^{(m)},y^{(m)})$
Note:
Mapping $x^{(i)} \to z^{(i)}$ should be defined by running PCA only on the training set. This mapping can be applied as well to the examples $x_{cv}^{(i)}$ and $x_{test}^{(i)}$ in the cross validation and test sets.
Application of PCA
- Compression
- Reduce memory/disk needed to store data
- Speed up learning algorithm
- Visualization
Exercise
- 课程原实验包括:pca可视化;用于人脸数据降维、重建
pca.py
# -*- coding: utf-8 -*-
"""
Created on Mon Jul 30 20:41:36 2018
@author: 周宝航
"""
import numpy as np
import numpy.linalg as la
class PCA(object):
def __init__(self):
pass
def featureNormalize(self, X):
m, n = X.shape
X_norm = X;
mu = np.zeros((1, n))
sigma = np.zeros((1, n))
for i in range(n):
mu[0, i] = np.mean(X[:,i])
sigma[0, i] = np.std(X[:,i])
X_norm = (X - mu) / sigma
return X_norm,mu,sigma
def pca(self, X):
m, n = X.shape
X_norm = X.T.dot(X) / m
U,S,_ = la.svd(X_norm)
return U,S
def projectData(self, X, U, K):
U_reduce = U[:, :K]
Z = X.dot(U_reduce)
return Z
def recoverData(self, Z, U, K):
U_reduce = U[:, :K]
X = Z.dot(U_reduce.T)
return X
main.py
# -*- coding: utf-8 -*-
"""
Created on Mon Jul 30 20:44:41 2018
@author: 周宝航
"""
from pca import PCA
import numpy as np
import scipy.io as sio
import matplotlib.pyplot as plt
# ================== Load Example Dataset ===================
data = sio.loadmat('data\\ex7data1.mat')
X = data['X']
m, n = X.shape
fig = plt.figure()
ax = fig.add_subplot(1,2,1)
ax.plot(X[:, 0], X[:, 1], 'bo')
# =============== Principal Component Analysis ===============
pca = PCA()
X_norm, mu, sigma = pca.featureNormalize(X)
U, S = pca.pca(X_norm)
p1 = mu
p2 = mu + 1.5 * S[0] * U[:, 0].T
data = np.r_[p1, p2].reshape([-1, 2])
ax.plot(data[:, 0], data[:, 1], '-k', linewidth=2)
p2 = mu + 1.5 * S[1] * U[:, 1].T
data = np.r_[p1, p2].reshape([-1, 2])
ax.plot(data[:, 0], data[:, 1], '-k', linewidth=2)
# =================== Dimension Reduction ===================
ax = fig.add_subplot(1,2,2)
ax.plot(X_norm[:, 0], X_norm[:, 1], 'bo')
K = 1
Z = pca.projectData(X_norm, U, K)
X_rec = pca.recoverData(Z, U, K)
ax.plot(X_rec[:, 0], X_rec[:, 1], 'ro')
for i in range(m):
data = np.r_[X_norm[i, :], X_rec[i, :]].reshape([-1, 2])
ax.plot(data[:, 0], data[:, 1], '--k', linewidth=2)
# =============== Loading and Visualizing Face Data =============
def displayData(X):
m, n = X.shape
example_width = np.int(np.sqrt(n))
example_height = np.int(n / example_width)
display_rows = np.int(np.floor(np.sqrt(m)))
display_cols = np.int(np.ceil(m / display_rows))
pad = 1;
display_array = - np.ones([pad + display_rows * (example_height + pad),\
pad + display_cols * (example_width + pad)])
curr_ex = 0
for j in range(display_rows):
for i in range(display_cols):
if curr_ex == m:
break
max_val = max(abs(X[curr_ex, :]))
row = pad+j*(example_height + pad)+1
col = pad+i*(example_width + pad)+1
display_array[row:row+example_height, col:col+example_width] = \
X[curr_ex, :].reshape([example_height, example_width]) / max_val
curr_ex += 1
if curr_ex == m:
break
plt.imshow(display_array.T, cmap='gray')
data = sio.loadmat('data\\ex7faces.mat')
X = data['X']
#displayData(X[1:100, :])
# =========== PCA on Face Data: Eigenfaces ===================
X_norm, mu, sigma = pca.featureNormalize(X)
U, S = pca.pca(X_norm)
#displayData(U[:, 1:36].T)
K = 100
Z = pca.projectData(X_norm, U, K)
# ==== Visualization of Faces after PCA Dimension Reduction ====
X_rec = pca.recoverData(Z, U, K)
fig = plt.figure()
original = fig.add_subplot(1, 2, 1)
original.set_title('Original faces')
displayData(X_norm[1:100,:])
recovered = fig.add_subplot(1, 2, 2)
recovered.set_title('Recovered faces')
displayData(X_rec[1:100,:])
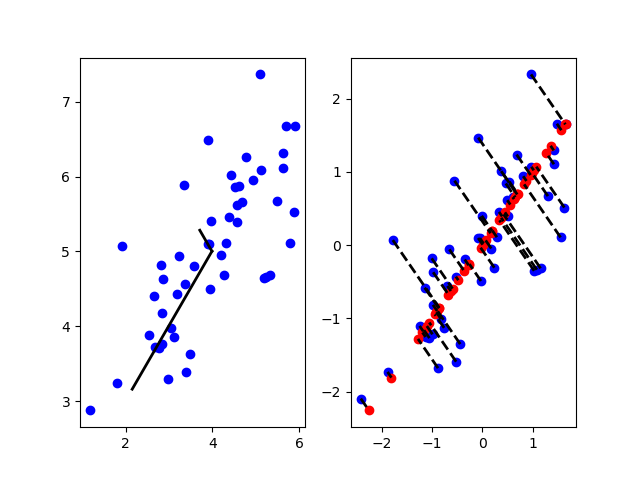
结果1
- 左图代表数据经过PCA处理得到的主成分方向。
- 右图中,蓝点表示原始数据、红点表示降维后的数据。虚线表示原始数据到降维后数据的映射。
结果2
- 左图为原始人脸图像数据,而右侧为降维后恢复的人脸数据。
